The science behind creating an accurate molecular representation of SARS-CoV-2 infection
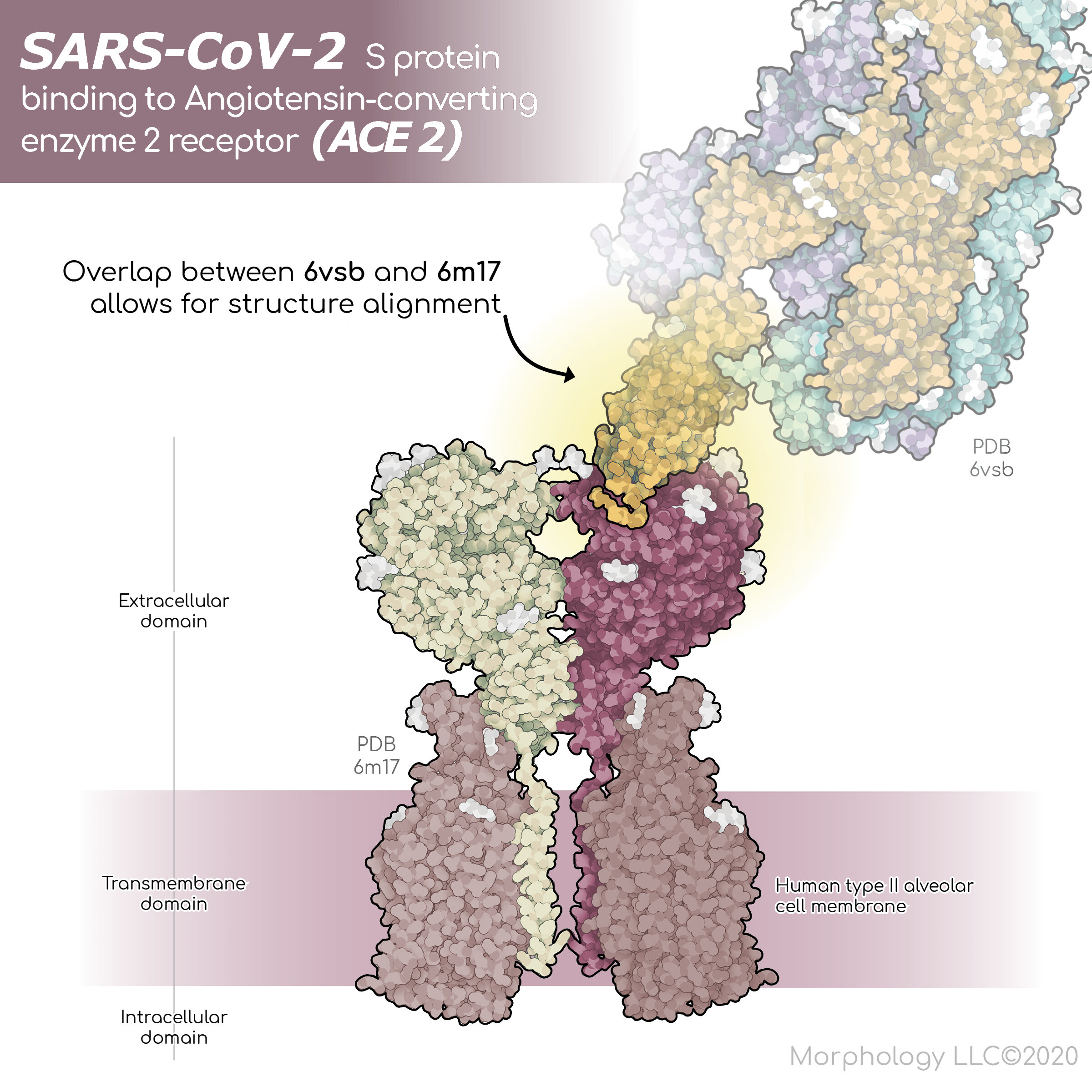
New cover art by Morphology Creative Director, Isabel Romero Calvo, MS, PhD, illustrates a scientifically accurate 3D model of SARS-CoV-2 infection. The image shows binding of the surface spike glycoprotein of the SARS-CoV-2 virus to an angiotensin-converting enzyme 2 receptor (ACES2) on a human type II alveolar cell, the cell type that lines the air sacs within the lungs. The binding of spike proteins to the receptor of the target cell is the first step of viral infection.
For research scientists, it is critical to use precise models for drug and vaccine development, and for molecular illustrators, it is necessary to understand how to accurately visualize these structures.
“In the world of molecular illustration, we produce works that are both science and art,” Isabel reflected. “In this way, we can view and interact with the structural components as if they were objects that we could hold in our hands, and maybe better understand them.”
Let’s dissect this cover art with Isabel, and learn about her process, including 1. tools she used to create these models, and 2. challenges and how to troubleshoot them.
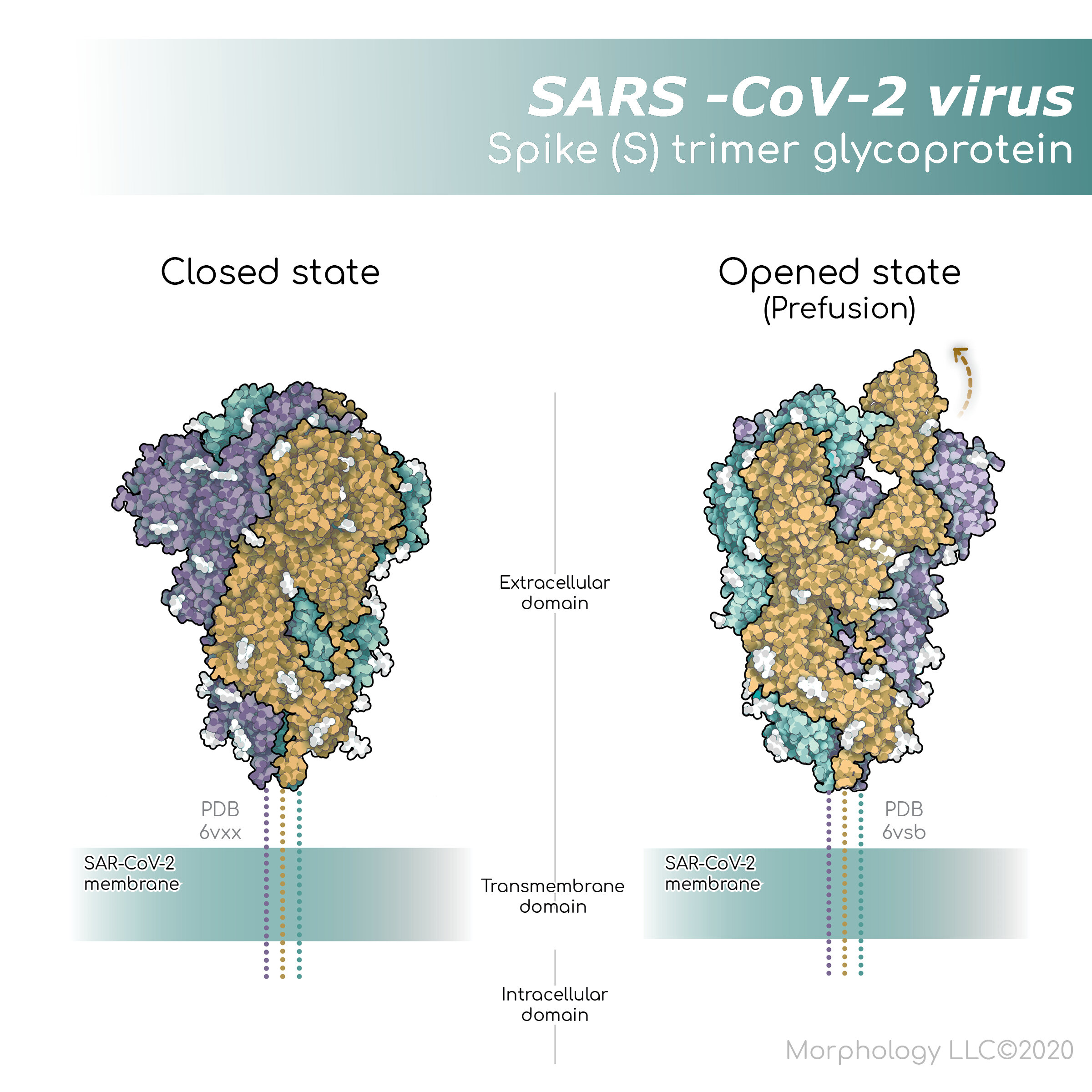
The SARS-CoV-2 virus spike (S) glycoprotein
The SARS-CoV-2 spike (S) protein is the largest protein on the viral surface and makes the first contact with a target cell. The spike protein is composed of three monomers, or three of the same peptides, which are entwined to form a single functional unit. It is a transmembrane protein; it has an intracellular, a transmembrane, and an extracellular domain, allowing it to sit securely within the viral membrane. The outside portion, or extracellular domain, interacts with proteins on the surface of the target cell.
Start with the structures
Where do scientists and molecular illustrators get accurate files of the amino acid sequences and structures of these proteins? They go to the Protein Data Bank (PDB), a data archive maintained by experts in structural and computational biology, that provides access to 3D structure data for large molecules, including proteins, DNA, and RNA. Visualizing the SARS-CoV-2 binding to ACE2, required structures of the spike glycoprotein of the SARS-CoV-2 virus (PDB 6VXX and 6VSB), and the angiotensin-converting enzyme 2 receptor (PDB 6M17).
Troubleshoot the challenges
Both amino acid sequence data, and crystallography structural data can be found in PDB, but that is far from the end of the story. It is critical to understand what is in these files because it’s not always straightforward. For some molecules, even if we know the peptide sequence, techniques used to reveal the structure may not be able to see the entire molecule. This can happen if sections of a molecule are flexible and dynamic. Typically, flexible regions with undefined structures are represented by dotted lines. Look closely at the spike S protein figure and you will notice dotted lines in the extracellular domain close to the viral membrane.
Another thing to keep in mind when thinking about structure is conformational changes. Proteins can and usually do change shape as they perform their jobs. For example, the spike protein of SARS-CoV-2 actually opens and closes to tightly grasp the receptor on the human cells. PDB contains a closed conformation (PDB 6VXX) (left panel), and an open conformation (PDB 6VSB) (right panel) of the S protein structure.
SARS-CoV-2 S protein binding to angiotensin-converting enzyme 2 receptor (ACE2)
The spike protein binds to angiotensin-converting enzyme 2 receptor (ACE2) (PDB 6M17). ACE2, like the viral spike protein, is transmembrane, meaning that is passes through the cell membrane. ACE2 may be found associated with other proteins in a tetramer complex consisting of four peptides.
Alignment tools
In case working with a tetramer complex isn’t tricky enough, the ACE2 PDB structural file contains sections of the viral spike protein. This detail is critical when creating a 3D scene. The ACE2 protein and the complete spike protein must be carefully overlayed to achieve an accurate representation. To accomplish this, the PDB structure files must be imported into a program called Visual Molecular Dynamics (VMD). The VMD MultiSeq tool can be used to complete the alignment.
In addition to protein alignment, it is important to place the transmembrane proteins perfectly within the cell membrane. Tools to assist with this task can be found in Orientations of Proteins in Membranes (OPM), and MemProtMD.
Note: The 2D molecular illustrations were created using Illustrate (Goodsell DS, Autin L, Olson AJ (2019) Illustrate: Software for Biomolecular Illustration. Structure 27, 1716-1720).
A diverse team of researchers
To add context to this cover design, Isabel integrated the molecular scene into a snapshot of daily work activity. In this way, she was able to incorporate the dedicated researchers into this visual story about responding to COVID-19.
To create this story, Isabel placed the 3D molecular composition into a contrasting 2D scene that could be a lab meeting or colleagues discussing science over a coffee. The 2D scene includes a tablet, surrounded by 2D vector-graphic hands of a diverse group of researchers. We don’t see the faces of the investigators, but we can see that they are discussing the mechanisms of infection of the virus, and we are optimistic that they are coming up with the answers.
The message of this cover goes beyond just molecules. Although the researchers are represented only by hands, this cover was an opportunity to show a diverse team of researchers working together. The Skin color palette illustrates the various skin tones used to create the hand vector graphics.
Isabel commented, “it is really our responsibility as scientific illustrators to show diversity at every available opportunity, even in a molecular piece.” She continued, “this is something we should constantly be thinking about as we create.”